Keyword: Gene expression
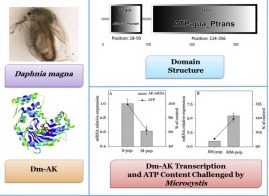
Lyu, K., L. Zhang, X. Zhu, G. Cui, A. E. Wilson, and Z. Yang. 2015. Arginine kinase in the cladoceran Daphnia magna: cDNA sequencing and expression is associated with resistance to toxic Microcystis. Aquatic Toxicology 160:13-21.
Abstract
Nutrient loading derived from anthropogenic activities into lakes have increased the frequency, severity and duration of toxic cyanobacterial blooms around the world. Although herbivorous zooplankton are generally considered to be unable to control toxic cyanobacteria, populations of some zooplankton, including Daphnia, have been shown to locally adapt to toxic cyanobacteria and suppress cyanobacterial bloom formation. However, little is known about the physiology of zooplankton behind this phenomenon. One possible explanation is that some zooplankton may induce more tolerance by elevating energy production, thereby adding more energy allocation to detoxification expenditure. It is assumed that arginine kinase (AK) serves as a core in temporal and spatial adenosine triphosphate (ATP) buffering in cells with high fluctuating energy requirements. To test this hypothesis, we studied the energetic response of a single Daphnia magna clone exposed to a toxic strain of Microcystis aeruginosa, PCC7806. Arginine kinase of D. magna (Dm-AK) was successfully cloned. An ATP-gua PtransN domain which was described as a guanidine substrate specificity domain and an ATP-gua Ptrans domain which was responsible for binding ATP were both identified in the Dm-AK. Phylogenetic analysis of AKs in a range of arthropod taxa suggested that Dm-AK was as dissimilar to other crustaceans as it was to insects. Dm-AK transcript level and ATP content in the presence of M. aeruginosa were significantly lower than those in the control diet containing only the nutritious chlorophyte, Scenedesmus obliquus, whereas the two parameters in the neonates whose mothers had been previously exposed to M. aeruginosa were significantly higher than those of mothers fed with pure S. obliquus. These findings suggest that Dm-AK might play an essential role in the coupling of energy production and utilization and the tolerance of D. magna to toxic cyanobacteria.
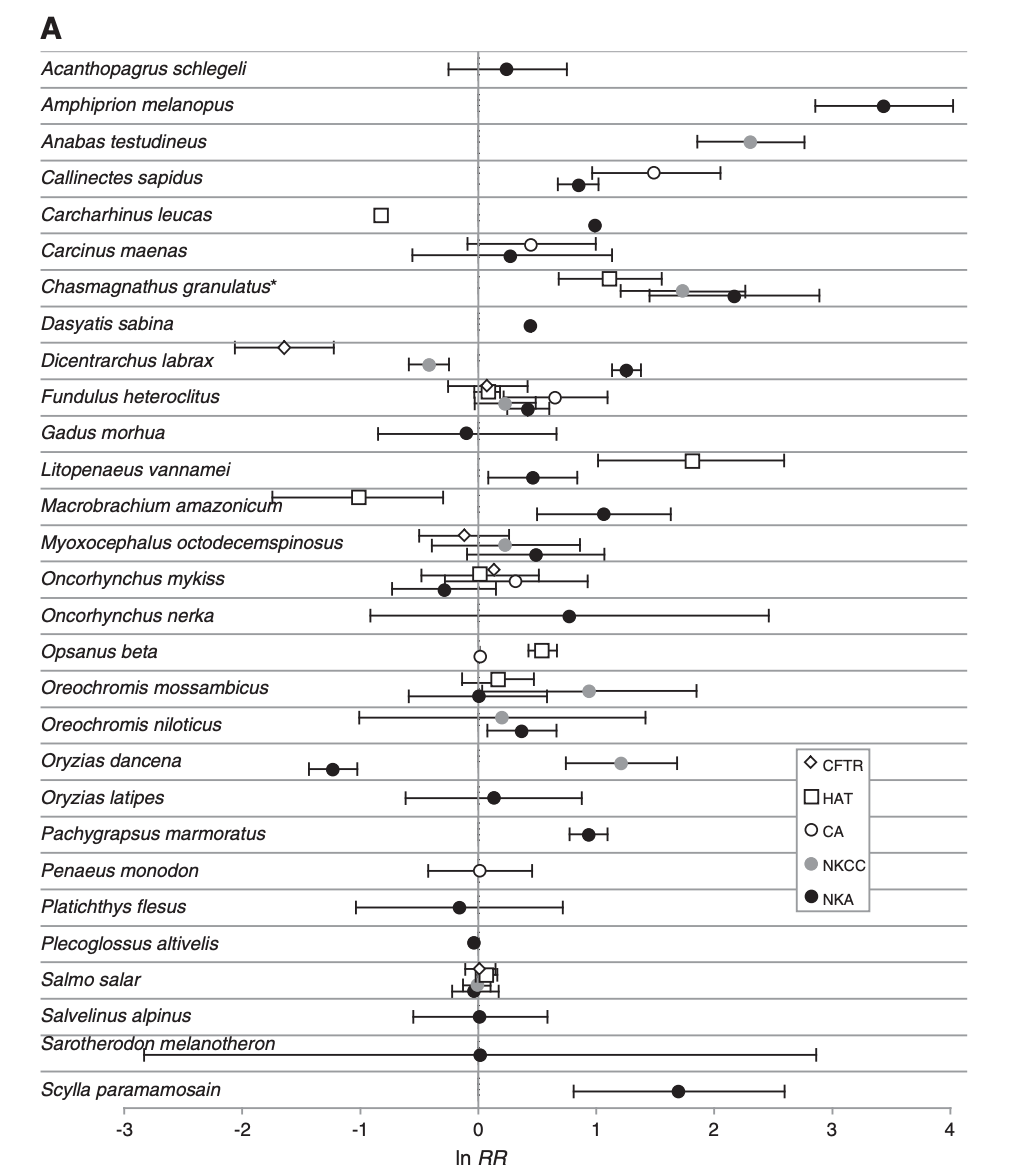
Havird, J. C., R. P. Henry, and A. E. Wilson. 2013. Altered expression of Na+/K+-ATPase and other osmoregulatory genes in the gills of euryhaline animals in response to salinity transfer: a meta-analysis of 59 quantitative PCR studies over 10 years. Comparative Biochemistry and Physiology, Part D: 8(2):131-140.
Abstract
Recent advances in molecular techniques have allowed gene expression in euryhaline animals to be quantified during salinity transfers. As these investigations transition from studying single genes to utilizing genomics-based methodologies, it is an appropriate time to summarize single gene studies. Therefore, a meta-analysis was performed on 59 published studies that used quantitative polymerase chain reaction (qPCR) to examine expression of osmoregulatory genes (the Na+/K+–ATPase, NKA; the Na+/K+/2Cl− cotransporter, NKCC; carbonic anhydrase, CA; the cystic fibrosis transmembrane regulator, CFTR; and the H+–ATPase, HAT) in response to salinity transfer. Based on 887 calculated effect sizes, NKA, NKCC, CA, and HAT are up-regulated after salinity transfer, while surprisingly, CFTR is unchanged. Meta-analysis also identified influential factors contributing to these changes. For example, expression was highest: 1) during transfers from higher to lower salinities comprising a physiological transition from osmoconformity to osmoregulation, 2) 1–3 days following transfer, 3) during dissimilar transfers, and 4) in crustaceans rather than teleosts. Methodological characteristics (e.g., types of controls) were not important. Experiments lacking in the current literature were also identified. Meta-analyses are powerful tools for quantitatively synthesizing a large body of literature, and this report serves as a template for their application in other areas of comparative physiology